News Story
BioE Researchers Develop Nano-guided Cell Networks as Conveyors of Molecular Communication

Fischell Department of Bioengineering (BioE) researchers have merged living cells with nanotechnology to develop an integrated molecular processing network that sheds light on how bioengineers might further bridge the communication gap between biology and electronic microfabricated devices.
In their Nature Communications paper published in mid-October, members of BioE Robert E. Fischell Distinguished Professor and Chair William Bentley’s research group addressed a question bioengineers have long contended with: how can scientists more effectively tap into the wealth of information that exists at the molecular level?
Advances in nanotechnology have provided bioengineers new ways to sample molecular space, but living cells have the capability to take things a step further in that they can identify molecules within complex environments and trigger functions.
When appropriately accessed, molecular information can provide bioengineers with invaluable feedback on biological systems, their componentry, and their functions.
To access this information, bioengineers have long worked to develop nano- to micro-scaled tools that engage with biological systems through monitoring and interacting at the molecular level. One such tool is synthetic biology, through which engineers “rewire” cells to survey molecular space – a feat that is possible because cells have sophisticated capabilities to recognize, amplify, and transduce chemical information.
Cells also “present a potential interface between chemically based biomolecular processing and conventional vectors of information flow, such as electrons and protons,” the group noted in Nature Communications.
Put simply, cells serve a crucial role as conveyors of molecular communication between biological systems, such as the gastrointestinal tract, and microdevices, such as stents.
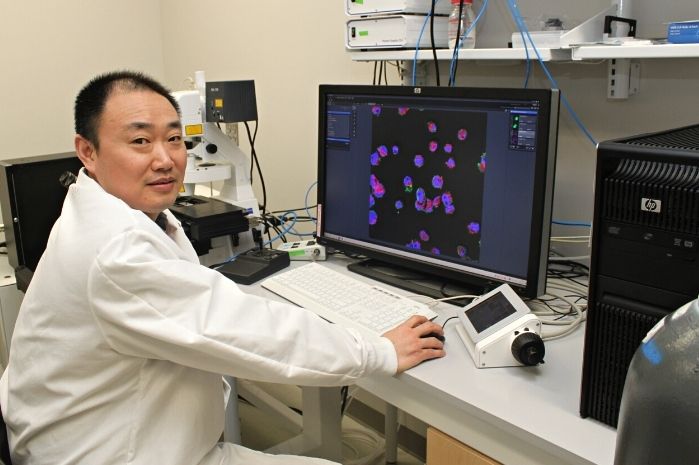
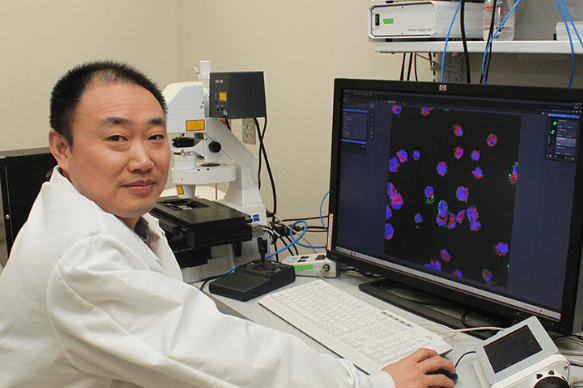
Building on this, Bentley’s research team developed a system in which a small network of surveyor cells collectively gathers information about the environment in which they live. Then, when they detect a target signal molecule, they synthesize “reporter” proteins onto their outer surface. One of the proteins is a fluorescent protein, the other facilitates binding to magnetic nanoparticles. In this way, these surveyor cells can be introduced to a particular biological niche and can then be collected with a magnet where their fluorescence will indicate the concentration of the analyte molecule they were engineered to find.
Eventually, this will enable “smart” bacteria that seek out pathogens or wounds that are revealed by molecule markers they emit. When the “smart bacteria” encounter pathogens or wounds, they synthesize and deliver a therapeutic at the right spot and the right time, and in the correct dose to counter the problem.
Bentley’s team consists of BioE/Institute for Bioscience and Biotechnology Research colleague Professor Gregory Payne, as well as collaborators Dr. John March (Associate Professor, Cornell University) and Dr. Matthew Chang (Associate Professor, National University of Singapore) and their research groups. The team's work is funded primarily by the U.S. Defense Threat Reduction Agency (DTRA).
BioE Ph.D. student Jessica Terrell, a member of Bentley’s Biomolecular and Metabolic Engineering Laboratories, served as lead author of the Nature Communications paper.
Published November 12, 2015