News Story
UMD Co-Leads Study That Could Aid Identification of Cell Defects
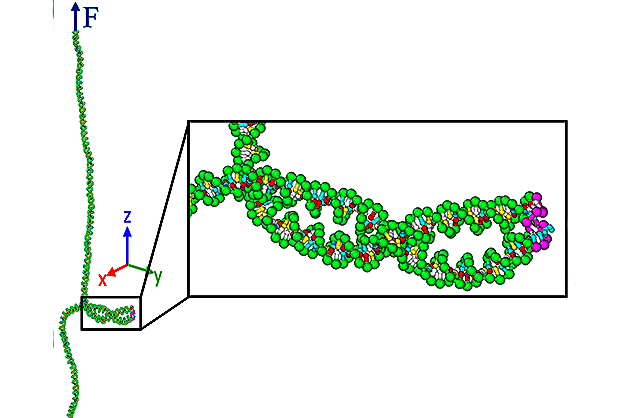
It’s sometimes compared to a rope ladder, albeit one that is twisted at intervals. The double-helix structure of DNA consists of two intertwining backbone strands, with chemical base pairs forming the “rungs.” On rare occasions—about one per ten million replications—a rung proves defective.
Now, researchers at the University of Maryland’s A. James Clark School of Engineering, in partnership with the National Institutes of Health (NIH) and researchers from Northwestern University and Rice University, have discovered a potential way to identify such genetic snafus more quickly.
The team, co-led by associate professor of mechanical engineering Siddhartha Das, used molecular dynamics simulations to study the relationship between cell defects—particularly mismatched DNA base pairs—and a phenomenon dubbed “supercoiling,” in which the sides of the “ladder” become more tightly twisted than usual, while the entire structure coils around itself. Dr. Keir Neuman of NIH’s National Heart Lungs Blood Institute is the other co-lead of this paper.
“We’ve found that in a supercoiled DNA, base pair mismatches are more likely to be found in the supercoil,” said UMD Ph.D. student Parth Rakesh Desai, a member of Das’s and Neuman’s research group. “This likelihood significantly reduces the number of base pairs that need to be checked in order to find a mismatch.” A paper detailing the team’s findings has now been published by Nucleic Acids Research, with Desai as the first author.
“Research of this kind not only provides an example of productive interdisciplinary collaboration but also illustrates the ways in which fields—in this case biology and mechanical engineering—can intersect, said Das, the UMD research group head.
 “We’re modeling one of the most important soft materials known to humanity, namely DNA,” he said. “It’s a logical extension of the other work we do in mechanical engineering, such as modeling polymers and other soft materials. The difference is that instead of applying these tools to manufacturing and other classical mechanical engineering problems, we’re applying it to a biological problem."
“We’re modeling one of the most important soft materials known to humanity, namely DNA,” he said. “It’s a logical extension of the other work we do in mechanical engineering, such as modeling polymers and other soft materials. The difference is that instead of applying these tools to manufacturing and other classical mechanical engineering problems, we’re applying it to a biological problem."
“We’re modeling one of the most important soft materials known to humanity, namely DNA.”
Dr. Siddhartha Das, associate professor of mechanical engineering
He stressed the role of the Graduate Partnership Program established by UMD and NIH’s National Heart Lungs Blood Institute (NHLBI) in supporting this vital research.
“The work is of great biophysical significance,” he said. “And we’ve been able to undertake it because of this partnership.” The work of Desai, the UMD Ph.D. student, has been funded through this program, Das noted.
UMD Mechanical Engineering Chair Bala Balachandran said the partnership with NHLBI was opening up new avenues for interdiscuplinary research. "We are excited to have this type of collaborative effort develop as a result of this partnership that was made possible by the spirited efforts of our Associate Chair for Research and Administration, Elkins Professor Don DeVoe," he said.
The team’s paper is titled “Coarse-Grained Modelling of DNA Plectoneme Pinning in the Presence of Base-Pair Mismatches.” In addition to Das, Desai, and Neuman, the authors include Sumitabha Brahmachari of Rice University and John F. Marko of Northwestern University.
Published November 2, 2020









